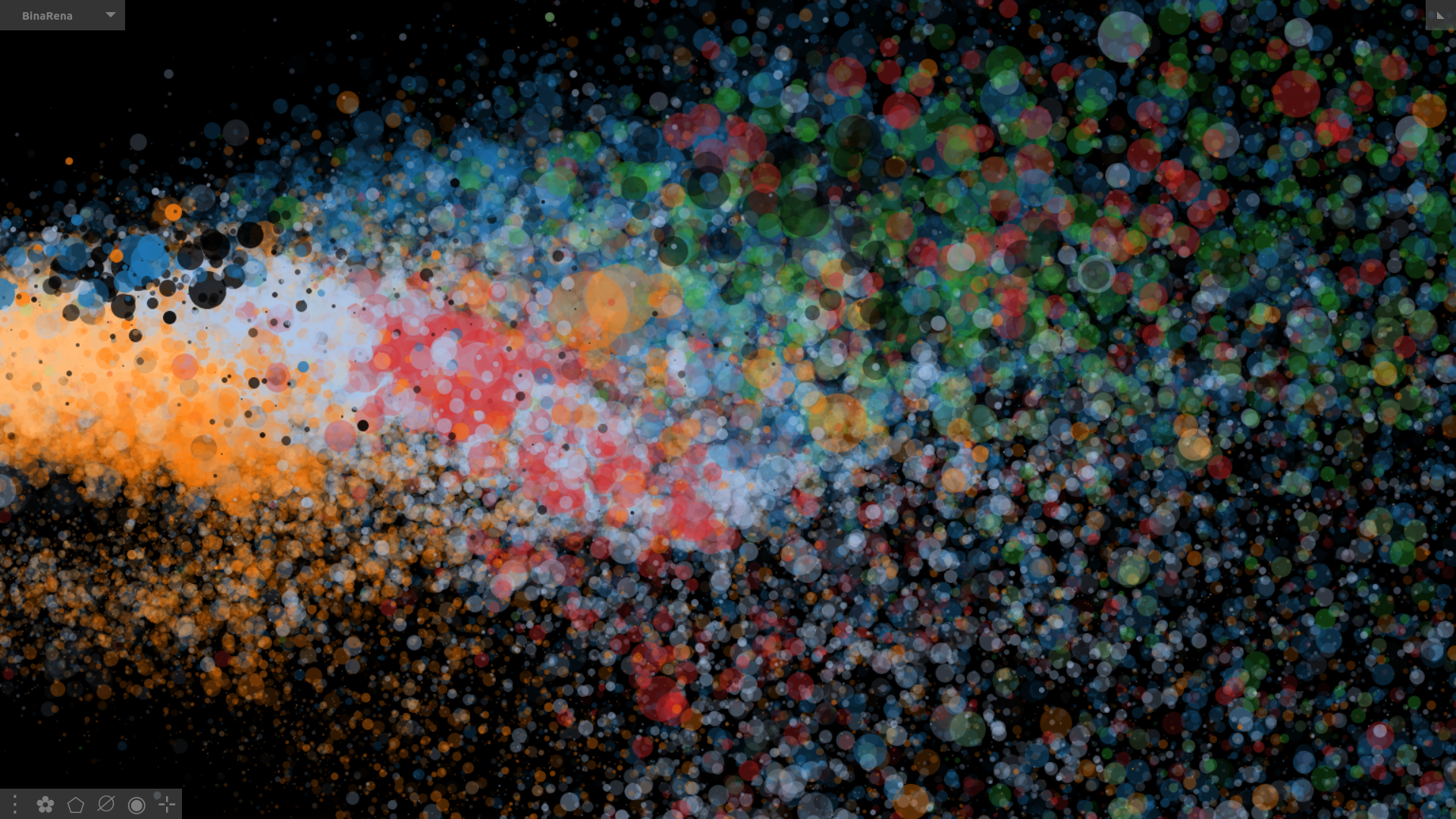Binarena ("bin arena") is an interactive visualizer that facilitates human-guided exploration and binning of metagenomic contigs.
Screenshot of BinaRena displaying a tropical peatland metagenome
Check out this live demo. It is a fully functional program with a pre-loaded sample dataset. Once you are familiar with the program, you may close the data, then drag and drop your own data into the interface and start to explore!
Download this program, unzip, and double-click "BinaRena.html". That's it!
BinaRena is a client-end web application. It is written in vanilla JavaScript, without using any third-party library. it does not require installation, nor does it require a web server running in the backend. In other words, it is literally a single webpage running in your browser.
- Dedicated to human operations. Designed to maximize comfort and productivity.
- For pattern discovery and hypothesis generation. For de novo binning and bin plan curation.
- Customizable plot visualizing various data types. Convenient manipulation of contigs and bins.
- Aesthetics, controls, reports and calculations integrated in UI. You get what you see.
The BinaRena Wiki provides comprehensive documentation of the program.
Quick links: Tutorial | Interface | Input | Plot | Contigs | Binning | Analysis | Output | Scripts | FAQ
The BinaRena program and its use cases in microbiome research have been described in the following paper:
MJ Pavia, A Chede, Z Wu, H Cadillo-Quiroz, Q Zhu. BinaRena: a dedicated interactive platform for human-guided exploration and binning of metagenomes. Microbiome. 2023. 11(1):186.
Please forward any questions to the project leader: Dr. Qiyun Zhu (qiyunzhu@gmail.com).