Releases: kjolley/BIGSdb
Version 1.46.2
BingMaps has been removed as an option for mapping as Microsoft will be retiring this service in June 2025. It has been replaced with options to use MapTiler (satellite) and ArcGIS streetmap/satellite imagery, alongside the existing option of OpenStreetMap for street level mapping.
OpenStreetMap
MapTiler
Esri/ArcGIS streetmap
Esri/ArcGIS satellite
You can select the appropriate options in bigsdb.conf (these can all also be overridden in individual database config.xml files if you wish to use different options or API keys):
#MAPPING############
#Set the option for displaying location maps.
#Note for Esri/ArcGIS services you must register
#[https://developers.arcgis.com/en/sign-up/]
#and abide by terms of service [https://developers.arcgis.com/en/terms/].
#For MapTiler you need to register and get an API key [https://www.maptiler.com/].
#0: OpenStreetMaps only
#1: OpenStreetMaps + MapTiler satellite imagery
#2: OpenStreetMaps + Esri/ArcGIS World Imagery (satellite)
#3: Esri/ArcGIS World Street Map + Esri/ArcGIS World Imagery (satellite)
mapping_option=0
;maptiler_api_key=xxxxxxxxxx
Note that use of MapTiler requires an API key - the free tier allows up to 100,000 map tile requests per month. You must also register for an Esri account to use Esri/ArcGIS services (you will need a paid account if using this commercially).
See the database setup documentation if you wish to enable mapping - either using a location field that contains GPS coordinates, or a geographic field such as 'town_or_city' which can be linked to a lookup table of GPS coordinates. The latter lookup table can be automatically populated to find the coordinates of towns and cities using the gp_town_lookups.pl script.
Full Changelog: v_1.46.1...v_1.46.2
Version 1.46.1
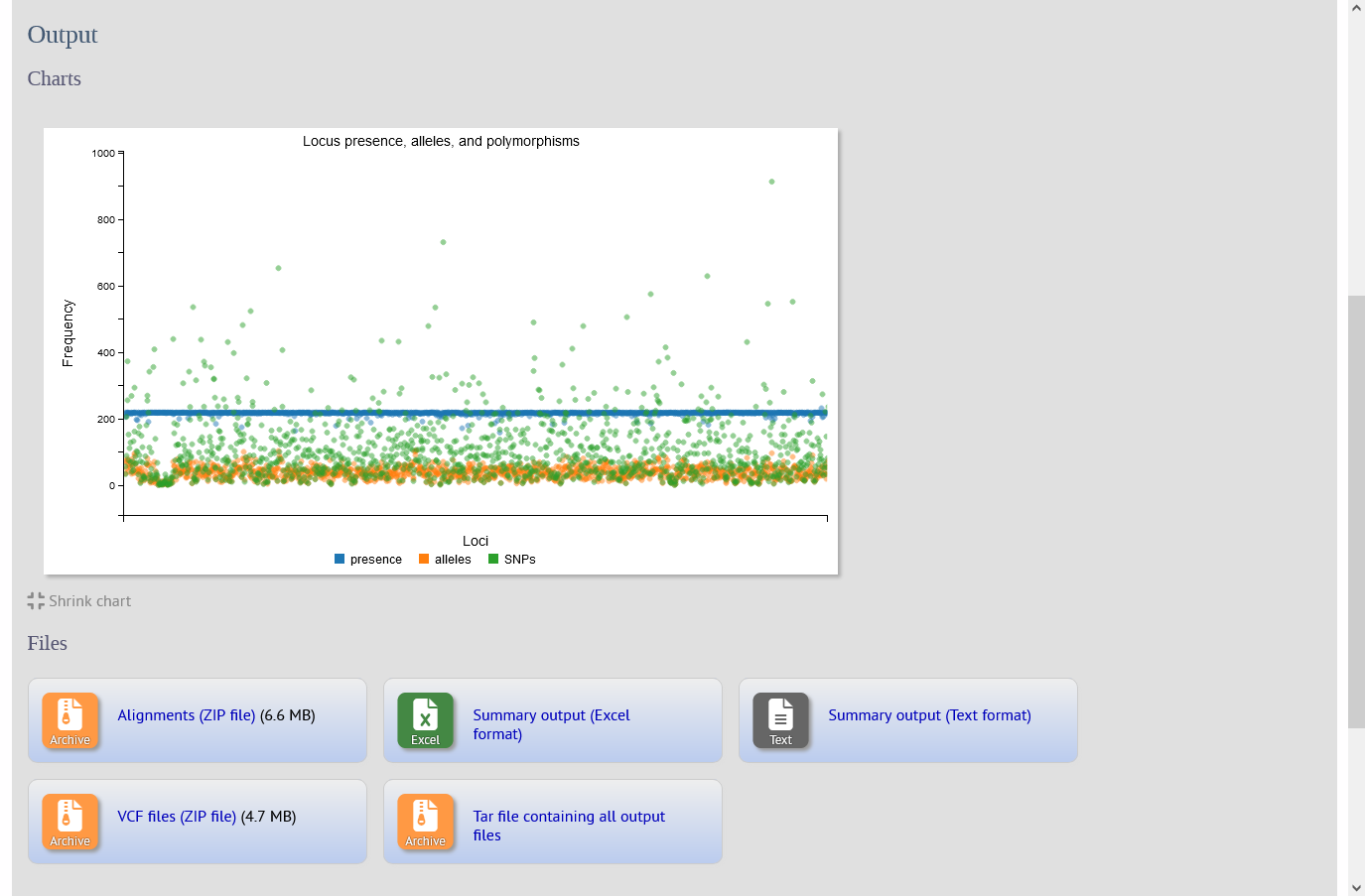
Version 1.46.1 introduces a new SNPsites plugin.
The SNPsites plugin aligns sequences for specified loci for an isolate dataset. The alignment is then passed to snp-sites to identify SNP positions. Output consists of a summary table including the number of alleles and polymorphic sites found for each locus, an interactive D3 chart that displays the summary, and ZIP files containing alignment FASTAs and VCF files for each locus.
Note that snp-sites needs to be installed and its path set in bigsdb.conf, e.g.
#snp-sites##########
snp_sites_path=/usr/bin/snp-sites
Full Changelog: v_1.46.0...v_1.46.1
Version 1.46.0
Version 1.46.0 introduces the following changes:
- An option has been added to filter alleles, profiles, and isolates for non-logged in users to records added before a specified date. This requires users to log in if they wish to access more recent data and has been introduced to provide licensing options. This can be activated globally in bigsdb.conf or within individual database config.xml files by setting the login_to_show_after_date attribute.
In bigsdb.conf:
login_to_show_after_date=2024-01-01
In config.xml:
login_to_show_after_date="2024-01-01"
- Fix for missing foreign key on the query_interface_fields table and missing primary key on scheme completion cache tables.
Full Changelog: v_1.45.0...v_1.46.0
Version 1.45.0
Version 1.45.0 introduces the following changes:
- An option has been added to highlight private data in the Excel file generated for isolate exports. If a user has access to private data (which may belong to others and been shared with them) there will be a section on the form that allows the user to select how these records are shaded within the output.
- An option has been added for an admin to create customized isolate query forms with pre-selected fields shown. New options will appear in the admin interface (with the Misc toggle selected):
Clicking the Add Query Interface link will allow a user to define new link text that will appear within the Search list on the contents page. Once this has been done, the Query Interface Fields links will become visible and this allows the admin to select any number of provenance, scheme, or LINcode fields to be pre-selected when the link is clicked.
- Finally, separate options have been added to force users to be logged in to download alleles or profiles when using the web interface. This is to encourage users to script downloads using the API. There are global settings in the bigsdb.conf file:
###DOWNLOADS################
#Require users to be logged in to download records via web interface
#Note that this is likely to prevent automated downloads (which generally
#should be done via API). These values can be overridden in the database
#config.xml file.
allele_downloads_require_login=1
profile_downloads_require_login=1
These can also be overridden on a per-database basis in the database config.xml file, e.g.
allele_downloads_require_login="yes"
profile_downloads_require_login="no"
Full Changelog: v_1.44.1...v_1.45.0
Version 1.44.1
Version 1.44.1 introduces the following changes:
- An option has been added to show if assembly checks have passed in the isolate results table. This can be enabled by default by setting
display_assembly_checks="yes"in the database config.xml file. It can also be set by end users in the user customization panels (https://bigsdb.readthedocs.io/en/latest/data_query/0100_options.html#main-results-table).
- An option has been added to scannew.pl to only scan records that have passed assembly checks.
- An option has also been added to not scan private records.
See https://bigsdb.readthedocs.io/en/latest/offline_tools.html#automated-offline-allele-definition for all scannew.pl options.
Full Changelog: v_1.44.0...v_1.44.1
Version 1.44.0
Version 1.44.0 introduces the following changes:
- New database indexes have been added to improve performance (thanks @braffes).
- Some rarely or never used indexes have been dropped.
- The option to set an allele designation with a status of 'ignore' has been removed. This was not really used but added an unnecessary filtering contraint on multiple queries.
- The number of isolates that match the current isolate at different LIN code thresholds are now displayed on an isolate record page (if LIN code has been assigned). The number is hyperlinked and takes you to a query that returns the matching isolates.
An administrator can also decide whether or not to include classification scheme or LIN code scheme matches on the isolate information page. These are enabled by default, if defined, but can be disabled with the following options in config.xml:
show_lincode_matches="no"
show_classification_schemes="no"
In addition, the number of LIN code thresholds to display can be set with the following option (replace the number with the required number of thresholds):
show_lincode_thresholds="5"
Full Changelog: v_1.43.3...v_1.44.0
Version 1.43.3
Version 1.43.3 brings the following changes:
- An optional message can be added on the allele and profile download pages to indicate that users should use the API for scripted data downloads. It is much easier to rate-limit the API than the web interface and it generally uses less resources to serve data. This message can be enabled by setting the URL for the API in bigsdb.conf, e.g.
rest_url=https://rest.pubmlst.org
- The JQuery.UI.multiselect widget is now used for dropdown boxes with large numbers of options, e.g. locus lists on the isolate query page, or where multiple options can be selected, such as the the 'includes' field list on various plugins. Where appropriate, the list is also searchable to make finding and selecting values quicker and easier.
- Javascript performance has been improved for background loading of isolate query form elements.
- Limits can now be set for the number of offline jobs that any single user can run for individual plugins. See the updated format of the job_limits.conf file found in the conf directory. If the limit is reached then further jobs for that particular plugin and user will be queued.
Full Changelog: v_1.43.2...v_1.43.3
Version 1.43.2
Version 1.43.2 brings the following changes:
- Additional logging information added. This includes the particular locus or scheme selected in data downloads, or the isolate record being shown.
- The cgST with fewest missing loci is now highlighted on the isolate info page when the isolate has multiple cgSTs.
- The query dashboard is now shown for bookmarked and publication queries.
- There is now an option to force users to log-in if they want to run offline jobs (see
jobs_require_login=1in bigsdb.conf). - There is now an option to automatically check the box to tag incomplete loci at specified identity thresholds (rather than just at 100% identity)
- 'Ambiguous read' flags are now automatically set on tags if sequence contains non-A,G,T,C characters.
- JQuery.UI.multiselect is now used for the flag dropdown box on the tag scan page. Previously we used an unformatted scrolling list box where an allele already had a flag but this was 4-5 lines long and made the table ugly and multiple flags difficult to select. For new tags there was a standard dropdown box which meant only one flag could be selected. As scan tables can be many thousands of rows long, the dropbox is only rendered when it is first visible in the current viewport.
Full Changelog: v_1.43.1...v_1.43.2
Version 1.43.1
Hotfix release to fix the isolate query modification trigger not appearing on the isolate query page (broken in commit d4b5439).
Full Changelog: v_1.43.0...v_1.43.1
Version 1.43.0
This version adds optional web logging to record user access. A new table log has been added to the bigsdb_auth database that records IP address, username, and page called with a timestamp. In addition, the REST API logging to the bigsdb_rest database has been improved by the addition of client and username information for when authenticated access is used.
To enable logging, you need to set the following in bigsdb.conf:
web_log_to_db=1
rest_log_to_db=1
Logging requires writing to the database on each page access so there is a very small performance penalty to enabling this. The tables are however unlogged (i.e. data are not written to the PosgreSQL write-ahead log) which makes them considerably faster than ordinary tables but data in them will be lost in the event of a database crash or unclean shutdown.
As every page access is recorded the log tables will grow in size over time. It is recommended that they are pruned regularly to remove records older than a specified period of time - this may also be required by GDPR! The easiest way to do this is to set up a scheduled CRON job by adding the following to /etc/crontab:
0 * * * * postgres psql -c "DELETE FROM log WHERE timestamp < NOW() - INTERVAL '7 days'" bigsdb_rest > /dev/null
10 * * * * postgres psql -c "DELETE FROM log WHERE timestamp < NOW() - INTERVAL '7 days'" bigsdb_auth > /dev/null
Additionally, there is a fix to the locus stats function in the sequence typing database necessitated by the introduction of schemes that can include locus presence/absence in their definitions. You therefore need to update the bigsdb_rest, bigsdb_auth, and any sequence definition database using the rest_v1.43.sql, auth_v1.43.sql, and seqdefdb_v1.43.sql scripts respectively. See https://github.com/kjolley/BIGSdb/blob/develop/Upgrading_README.txt.
Note that in the unlikely event that you have defined schemes that utilize locus presence/absence (where profiles can include the 'P' designation to indicate 'presence'), rather than just sequence variation, you should update the locus stats within the typing database with the following commands, otherwise the reported minimum allele length may be wrong.
DELETE FROM locus_stats;
INSERT INTO locus_stats(locus,datestamp,allele_count,min_length,max_length)
SELECT loci.id,MAX(sequences.datestamp),COUNT(sequences.allele_id),MIN(LENGTH(sequence)),MAX(LENGTH(sequence))
FROM loci LEFT JOIN sequences ON loci.id=sequences.locus
WHERE allele_id NOT IN ('N','0','P') OR allele_id IS NULL
GROUP BY loci.id;
Full Changelog: v_1.42.7...v_1.43.0