-
Notifications
You must be signed in to change notification settings - Fork 174
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Off-by-one in LinearFilter.General step #938
Comments
|
Actually I think there's something slightly more serious/general going on, with how the discretization has been incorrectly formulated to be preemptive with the state at the cost of effectively predicting the future passthrough. This basically means that transfer functions that are not strictly proper are not possibly going to be able to behave correctly (and fortunately Lowpass/Alpha are strictly proper, so they have no passthrough). So, I need to be more sure of what this means should happen in Nengo, so I'm closing this issue for now. Sorry for the spam. |
|
I think I've found the root of the problem. If we put an infinitesimally small passthrough on the system (which keeps the system's intended behaviour identical), then suddenly things change because Nengo inserts a delay. The blue line in the second graph is Nengo, which is inconsistent from the first simulation. The green lines are from I believe the issue revolves around this line Changing |
|
I think this is fixed in #1535. Here's the plots I get for the first systems: |
|
If we use the example from #1535 (comment) or For For import pylab
import numpy as np
from scipy.signal import lfilter, cont2discrete, tf2ss
import nengo
sys1 = nengo.LinearFilter([-1, 0, 1e6], [1, 2, 1])
sys2 = nengo.LinearFilter([0.0001, 0, 0], [0.0001, 0.02, 1])
for sys in (sys1, sys2):
with nengo.Network() as model:
x = nengo.Node(output=nengo.processes.PresentInput([1000, 0], 0.001))
p = nengo.Probe(x, synapse=sys)
p_x = nengo.Probe(x, synapse=None)
sim = nengo.Simulator(model)
sim.run(0.02)
(num,), den, _ = cont2discrete((sys.num, sys.den), sim.dt)
expected = lfilter(num, den, sim.data[p_x].squeeze())
pylab.figure()
pylab.title("len(num)=%d, len(den)=%d" % (len(sys.num), len(sys.den)))
pylab.plot(sim.trange(), sim.data[p].squeeze() - expected.squeeze())
#pylab.plot(sim.trange(), expected)
pylab.xlabel("Time (s)")
pylab.ylabel("Error")
pylab.show()I believe the reason is that the discrete state-space model for an LTI is defined by equation (5.5): which is also what nengolib does. However, #1535 uses the equations: Note the difference is in I believe the reason we are doing that is because there is already a delay elsewhere in Nengo. Prior to #1535 (and in nengolib) this delay was compensated for by applying the z-operator to the transfer function (which you can do if and only if it is strictly proper -- that is, no passthrough). This also meant that when In #1535, this is instead compensated for by using the most recent value of A correct fix would be to use In the context of #1535 I believe this means there won't be an ideal fix unless there is an easy way to eliminate the delay elsewhere in the graph, and move it explicitly into the synapse. Circular dependencies would occur if and only if all of the synapses along the chain have |
|
I spent a few hours and figured out the proper way to implement synapses when To follow my recommendation of "moving the delay explicitly into the synapse" we need to split up the The biggest issue with this approach is it requires special builder logic that only makes sense for linear transfer functions, but it makes a difference in behaviour (compared to #1535) by correcting the case of One possible advantage of this change (aside from correcting the behaviour) is it would give the operator optimizer the ability to group together all of the synapse multiplies (because it would be expressed in terms of only basic operators). I would expect this to give performance gains when there are lots of synapses between small ensembles. |
|
PR #1536 provides a proof-of-concept implementation of the above suggestion. |
|
One caveat with this is that many backends will not support this, particularly the neuromorphic ones. That said, e.g. Loihi doesn't support any synapses that can't be made out of exponential filters, so My impression is that |
Other backends (e.g., nengo-ocl) could keep doing the same thing that Nengo's backend did prior to #1535, and that was to have a two time-step delay per synapse whenever
One possibility is in modelling gap junctions (electrical synapses) when
Here are two quick neuromorphic use-cases:
However, neither transfer function does the correct thing with #1535, and produces an extra delay on master. #1536 solves both problems.
So in #1536, the situations where a recurrent synapse would work would be In my opinion it's much stranger to invent a different definition of an LTI system that does not implement the discrete as #1535 does (note that this achieves equality if and only if |
|
Here's another quick example to demonstrate some of the weirdness of implementing a different transfer function from what was requested. This applies to both master and #1535, but it is different than the other examples because it does not invoke the import nengo
sys1 = nengo.LinearFilter([1], [1, 0], analog=False)
sys2 = nengo.LinearFilter([1], [1], analog=False)
for sys in (sys1, sys2):
with nengo.Network() as model:
x = nengo.Node(output=nengo.processes.PresentInput([1000, 0], 0.001))
p = nengo.Probe(x, synapse=sys)
p_x = nengo.Probe(x, synapse=None)
with nengo.Simulator(model, progress_bar=False) as sim:
sim.run_steps(4, progress_bar=False)
print(sim.data[p].squeeze(), sys.filt(sim.data[p_x].squeeze(), y0=0))This example demonstrates two other forms of odd behaviour that stem from the above discussion:
#1536 solves both issues (tests have been modified and added corresponding to both points). |
I forgot to mention perhaps the earliest use-case for
This paper argues that the analog transfer function |



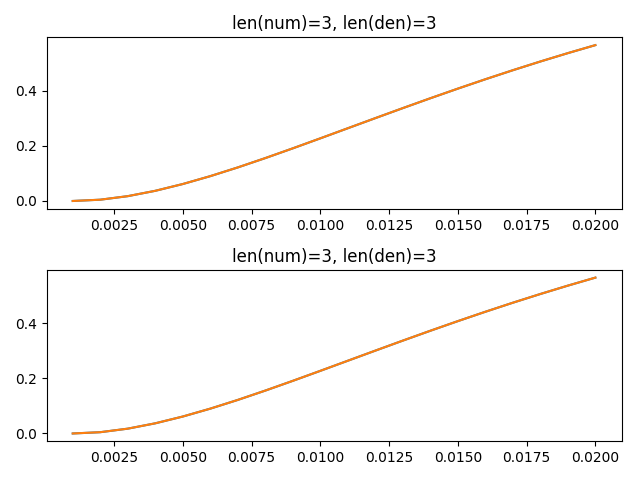
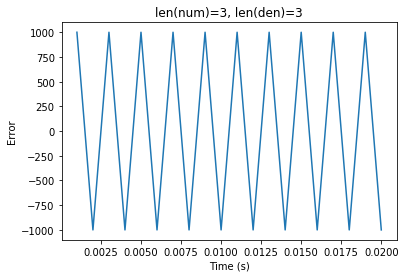
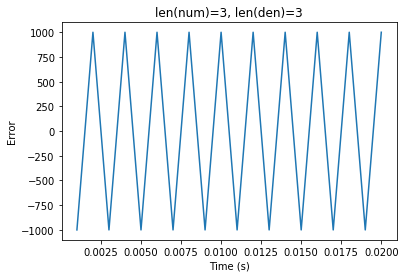
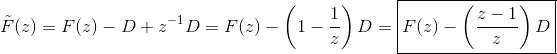
When simulating higher order synapses, the
Generalstep method is invoked, which seems to be off-by-one delay, possibly due to the ordering in whichydepends onoutput.The following test shows that
scipy.signal.lfilteragrees with the synapse implementation in theSimplestep for a first-order synapse such asLowpass, but not for a higher order differentiator.Currently, my implementation in NengoLib avoids this problem by simulating the state-space directly. Moreover, by putting it in controllable form, the
Amatrix reduces to a dot product and a shift operation. This requires less state than the LCCD method, and also reduces total simulation time.The text was updated successfully, but these errors were encountered: