-
-
Notifications
You must be signed in to change notification settings - Fork 54
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
extraction error #422
Comments
|
Hi @CaiYitao could you provide detailed info about:
|
|
Rocky Linux 8.6 Thank you for your prompt reply! |
|
Hi @CaiYitao . We will respond ASAP, probably next week. |
|
Hi @ernestoarbitrio, Could we extract all the parts in one slide now? |
|
Hi @CaiYitao
The result you're showing is compatible with the default tiling behavior, i.e., extracting tiles from the biggest tissue portion in the slide. This behavior is controlled by the parameter Therefore, if you want to extract tiles from all the tissue found in the slide you need to instantiate the from histolab.masks import TissueMask
# your code here
grid_tiles_extractor.locate_tiles(
slide=slide,
extraction_mask=TissueMask(),
# your params
) |
|
Hi @alessiamarcolini, Did we find out why it took a very long time to process the .mrxs file? BTW, can we write the tiles as jpg? when I set the suffix= ".jpg" and extract the tiles. Thank you! |
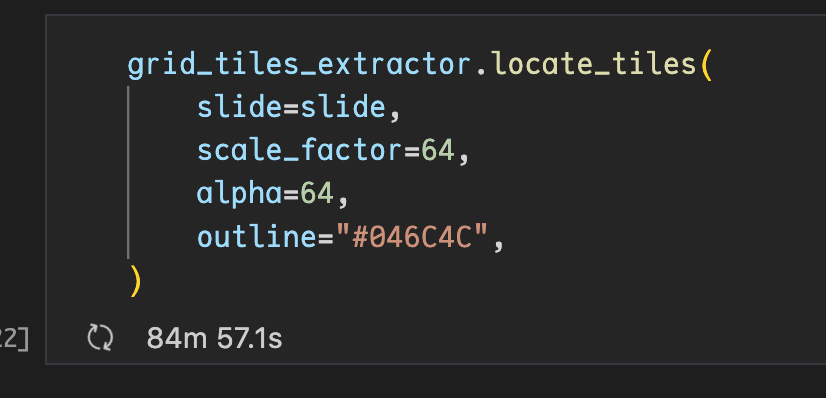





Discussed in #420
Originally posted by CaiYitao July 6, 2022
Just tried out to extract GridTiler, but get the following error:
Is it because my PIL library is not compatible with histolab?
my slide is in ".mrxs" format, I also tried to set suffix to .jpg but got same error
The text was updated successfully, but these errors were encountered: