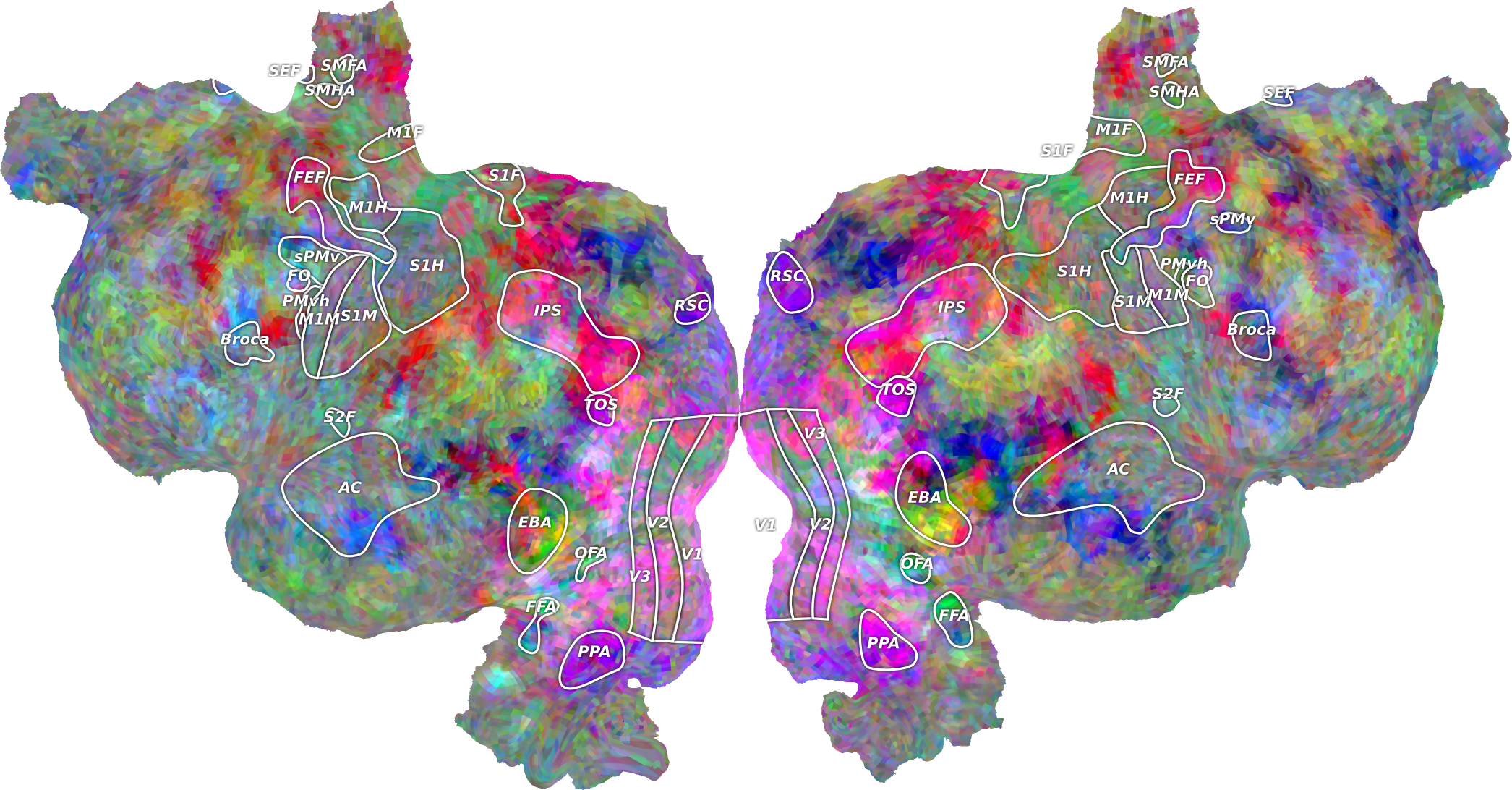
Pycortex is a software library that allows you to visualize fMRI or other volumetric neuroimaging data on cortical surfaces.
Note
Pycortex cannot be currently installed in Windows, only Linux and macOS are supported.
To install the stable release version of pycortex, do the following:
# First, install some required dependencies (if not already installed)
pip install -U setuptools wheel numpy cython
# Install the latest release of pycortex from pip
pip install -U pycortexIf you wish to install the development version of pycortex, you can install it directly from Github.
To do so, replace the second install line above with the following:
# Install development version of pycortex from github
pip install -U git+https://github.com/gallantlab/pycortex.git --no-build-isolationPycortex documentation is available at https://gallantlab.github.io/pycortex.
You can find many examples of pycortex features in the pycortex example gallery.
To build the documentation locally:
# Install required dependencies for the documentation
pip install sphinx_gallery numpydoc
# Move into the docs folder (assuming already in pycortex directory)
cd docs
# Build a local version of the documentation site
make htmlAfter you run the above, you can open docs/_build/html/index.html in a web browser to view the locally built documentation.
Pycortex is best used with IPython.
If you do not already have IPython, you can install it by running:
pip install ipythonTo run the pycortex demo, using IPython, run:
$ ipython
In [1]: import cortex
In [2]: cortex.webshow(cortex.Volume.random("S1", "fullhead"))
If you use pycortex in published work, please cite the pycortex paper:
Gao JS, Huth AG, Lescroart MD and Gallant JL (2015) Pycortex: an interactive surface visualizer for fMRI. Front. Neuroinform. 9:23. doi: 10.3389/fninf.2015.00023