-
Notifications
You must be signed in to change notification settings - Fork 30
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
adjust width based on density #51
Comments
|
It does seem reasonable to match produces matching violins and points. |
|
Yes - matching the default would be nice. Would it be possible to have |
|
I would like to also request a similar feature for For instance, you can see below that Thanks for your work on this useful package! library(ggbeeswarm)
#> Loading required package: ggplot2
dat <- structure(list(x = structure(c(1L, 1L, 2L, 1L, 1L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 1L, 1L,
2L, 1L, 1L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L,
2L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 2L, 1L), .Label = c("1",
"2"), class = "factor"),
y = c(0.00743015064770599, 0.00402350008901549,
0.0182015059430295, 0.0931013077022777, 0.0201296467502311, 0.00988912196583758,
0.00890949491496453, 0.0040521814310705, 0.0748063422432635,
0.0028265114618079, 0.0033580728734968, 0.0011570364882694, 0.00174334311680045,
0.300698451786883, 0.00186244761866073, 0.0117684502764761, 0.00578161092355426,
0.00651988531447871, 0.079985399989931, 0.00184351402134222,
0.00871624623132749, 0.00274545650752541, 0.00481711124028504,
0.00301568797492199, 0.00206841018634773, 0.00678981408558268,
0.00222290514238885, 0.00984868656785107, 0.00162418058758061,
0.00557005888481755, 0.121683319866249, 0.0030356653660198, 0.00210656116734316,
0.00353151727636117, 0.000903423206896699, 0.0162988509558787,
0.000853474507087549, 0.00220279909556151, 0.000414831444223472,
0.00233948134370524, 0.019787161002403, 0.00318015236414923,
0.00257190063774256, 0.0176909379302089, 0.00598338665216149,
0.136596738744799, 0.00128003795128495, 0.0202978906365311, 0.00156548169986173,
0.0168584387682024, 0.252985835349248)), row.names = c(NA, -51L
), class = c("tbl_df", "tbl", "data.frame"))
ggplot(dat, aes(x, y)) +
theme_bw(base_size = 22) +
geom_violin(draw_quantiles = c(0.5),
scale="width", width=0.6) +
geom_beeswarm(size=0.5, cex=1.5, priority = "random") +
ggtitle("cex = 1.5")
#> Warning in regularize.values(x, y, ties, missing(ties)): collapsing to unique
#> 'x' valuesggplot(dat, aes(x, y)) +
theme_bw(base_size = 22) +
geom_violin(draw_quantiles = c(0.5),
scale="width", width=0.6) +
geom_beeswarm(size=0.5, cex=2.5, priority = "random") +
ggtitle("cex = 2.5")
#> Warning in regularize.values(x, y, ties, missing(ties)): collapsing to unique
#> 'x' valuesCreated on 2020-09-08 by the reprex package (v0.3.0) |
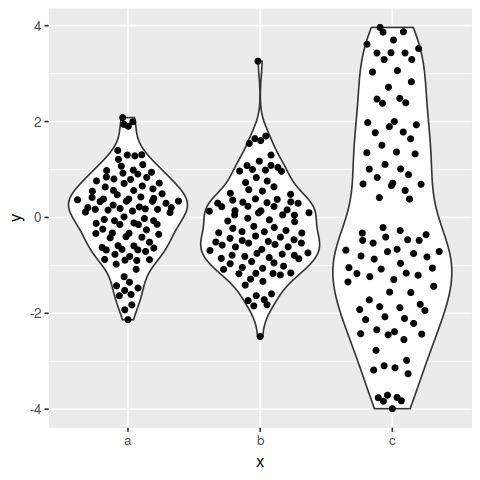


Hi - thanks for this package, it's really super!
Is it possible to adjust the width of the point distribution based on the density - similar to how geom_violin() works? I see that you can adjust the width based on the number of points, but it's not quite the same. I've given an example below - ideally, it would be possible to have the points inside the violin in the third column as well. Something analogous to
geom_violin(scale = 'area').Thank you,
Benjamin
The text was updated successfully, but these errors were encountered: